Promotional price valid on web orders only. Your contract pricing may differ. Interested in signing up for a dedicated account number?
Learn More
Learn More
Human KIR/CD158 Fluorescein-conjugated Antibody, R&D Systems™


Mouse Monoclonal Antibody has been used in 2 publications
Supplier: R&D Systems FAB1848F
This item is not returnable.
View return policy
Description
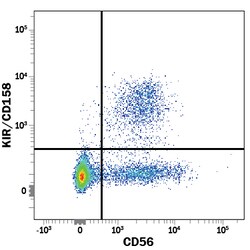
KIR/CD158 Monoclonal specifically detects KIR/CD158 in Human samples. It is validated for Flow Cytometry.Specifications
| KIR/CD158 | |
| Monoclonal | |
| Fluorescein | |
| Supplied in a saline solution containing BSA and Sodium Azide. with Sodium Azide | |
| KIR2DL1 | |
| BaF3 mouse pro-B cell line transfected with human KIR2DL3 | |
| 100 Tests | |
| Primary | |
| Detects human KIR/CD158. Stains cells transfected with KIR2DL2, 2DL3, 2DS2, or 2DS4. It does not stain cells transfected with KIR2DL1, 2DL4, 3DL1, or 3DL2. | |
| Protect from light. Do not freeze. 12 months from date of receipt, 2 to 8 degreesC as supplied. | |
| IgG2b |
| Flow Cytometry | |
| 180704 | |
| Flow Cytometry 10 uL/10^6 cells | |
| CD158, CD158 antigen-like family member A, CD158a, CD158a antigen, CD158Ankat1, CD158b1, CD158b2, CD158d, CD158e2, CD158f, CD158i, CD158k, cl-42, CL-43, CL-5, killer cell immunoglobulin-like receptor 2DL1, killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1, killer Ig receptor, killer inhibitory receptor 2-2-1, KIR221, KIR2DL5A, KIR-G1, MHC class I NK cell receptor, Natural killer-associated transcript 1, NKAT, NKAT-1, nkat10, NKAT1KIR-K64, NKAT2, NKAT4A, NKAT4B, NKAT6, p58 killer cell inhibitory receptor KIR-K64, p58 natural killer cell receptor clones CL-42/47.11, p58 NK cell inhibitory receptor NKR-K6,47.11, p58 NK receptor CL-42/47.11, p58.1, p58.1 MHC class-I-specific NK receptor | |
| Mouse | |
| Protein A or G purified from hybridoma culture supernatant | |
| RUO | |
| 3802 | |
| Human | |
| Purified |
Product Content Correction
Your input is important to us. Please complete this form to provide feedback related to the content on this product.
Product Title
Spot an opportunity for improvement?Share a Content Correction