Promotional price valid on web orders only. Your contract pricing may differ. Interested in signing up for a dedicated account number?
Learn More
Learn More
Invitrogen™ NCKAP1 Polyclonal Antibody
Rabbit Polyclonal Antibody
Supplier: Invitrogen™ PA530406
Description
Recommended positive controls: U87-MG. Predicted reactivity: Mouse (98%), Rat (97%), Zebrafish (92%), Xenopus laevis (94%), Rhesus Monkey (100%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
This gene interacts with Nck and may be involved in the downstream signaling of Nck and shares similarity with the mouse Dhm1 and the yeast dhp1 gene. The yeast gene is involved in homologous recombination and RNA metabolism, such as RNA synthesis and RNA trafficking. Complementation studies show that Dhm1 has a similar function in mouse as dhp1. The function of the human gene has not yet been determined. Transcript variants encoding different isoforms have been noted for this gene; however, their full-length nature is not known.
Specifications
| NCKAP1 | |
| Polyclonal | |
| Unconjugated | |
| NCKAP1 | |
| Brain protein H19; C79304; H19; HEM2; Hem-2; KIAA0587; Membrane-associated protein HEM-2; MH19; mKIAA0587; NAP 1; Nap1; NAP125; NCK associated protein 1; NCKAP1; NCK-associated protein 1; p125Nap1 | |
| Rabbit | |
| Antigen Affinity Chromatography | |
| RUO | |
| 10787, 50884 | |
| Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles. | |
| Liquid |
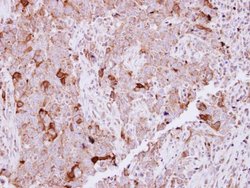
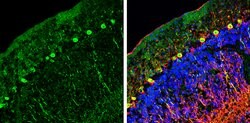
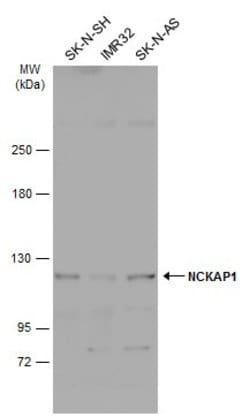
| Immunohistochemistry (Frozen), Immunohistochemistry (Paraffin), Western Blot | |
| 0.36 mg/mL | |
| PBS with 1% BSA, 20% glycerol and 0.025% ProClin 300; pH 7 | |
| P28660, Q9Y2A7 | |
| NCKAP1 | |
| Recombinant fragment corresponding to a region within amino acids 2 and 338 of Human NCKAP1. | |
| 100 μL | |
| Primary | |
| Human, Mouse | |
| Antibody | |
| IgG |
Safety and Handling
WARNING: Cancer - www.P65Warnings.ca.gov
Product Content Correction
Your input is important to us. Please complete this form to provide feedback related to the content on this product.
Product Title
Spot an opportunity for improvement?Share a Content Correction