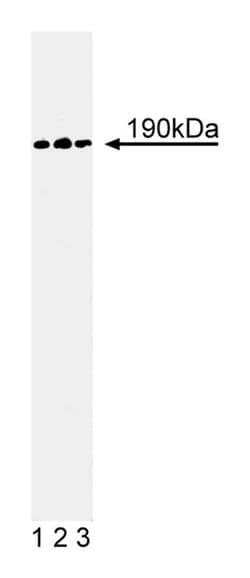Learn More
Description
GTPase activating proteins (GAPs) stimulate the GTP-hydrolyzing activity of GTPases, such as p21ras and Rho. p190-A is a Ras-GAP associated protein that is tyrosine phosphorylated in transformed and growth factor-stimulated cells. Ras-GAP and p190-A are targets of oncoproteins and growth factor receptors. p190-B is another Ras-GAP in the p190 family. It has 51% identity with p190-A and contains several GTPase-related domains in the N-terminal region and a Rho-GAP domain in the C-terminal region. p190-B is expressed in kidney, brain, liver, and lung, as well as in human foreskin fibroblasts, RD muscle cells, and HT-1080 cells. In fibroblasts, p190-B is localized diffusely in the cytoplasm and co-localizes with the α5β1 integrin receptor for fibronectin. Adhesion of fibronectin-coated latex beads to cells leads to the recruitment of p190-B and Rho to the plasma membrane at sites of bead contact. In addition, the recombinant Rho-GAP domain of p190-B displays GAP activity for RhoA, Rac1, and G25K/CDC42Hs. Thus, p190-B is thought to act as a transmembrane link between integrins and Rho GTPases during fibronectin-induced changes in cell morphology and motility.
Host Species: Mouse
Clone: 54
Isotype: IgG1
Species Reactivity [for Features Main]: Human
Immunogen: Human p190-B aa. 1102-1214
Immunofluorescence, Western Blotting

Specifications
Specifications
| Antigen | p190-B |
| Applications | Western Blot |
| Classification | Monoclonal |
| Clone | 54 |
| Concentration | 250μg/mL |
| Conjugate | Unconjugated |
| Formulation | Aqueous buffered solution containing BSA, glycerol, and ≤0.09% sodium azide. |
| Host Species | Mouse |
| Immunogen | Human p190-B aa. 1102-1214 |
| Purification Method | Affinity Purified |
| Show More |
For Research Use Only.
By clicking Submit, you acknowledge that you may be contacted by Fisher Scientific in regards to the feedback you have provided in this form. We will not share your information for any other purposes. All contact information provided shall also be maintained in accordance with our Privacy Policy.